Introduction
Millions of migratory and other birds are trapped, traded and consumed each year in most of the Mediterranean Basin, especially in the Middle East, North Africa and Malta (Brochet et al. Reference Brochet, Van Den Bossche, Jbour, Ndang’Ang’a, Jones, Abdou, Al-Hmoud, Asswad, Atienza, Atrash, Barbara, Bensusan, Bino, Celada, Cherkaoui, Costa, Deceuninck, Etayeb, Feltrup-Azafzaf, Figelj, Gustin, Kmecl, Kocevski, Korbeti, Kotrošan, Laguna, Lattuada, Leităo, Lopes, López-Jiménez, Lucić, Micol, Moali, Perlman, Piludu, Portolou, Putilin, Quaintenne, Ramadan-Jaradi, Ružić, Sandor, Sarajli, Saveljić, Sheldon, Shialis, Tsiopelas, Vargas, Thompson, Brunner, Grimmett and Butchart2016). Nevertheless, documentation of the actual extent of this activity and its possible effects on bird populations is scanty, partly as a consequence of its often-illegal nature that raises multiple obstacles to proper scientific study. Although regulation of legal hunting activities has been a central tenet of the environmental policies of all EU countries, the degree and extent of illegal bird killing has been assessed only very recently at the European level (Hirschfeld and Heyd Reference Hirschfeld and Heyd2005, Brochet et al. Reference Brochet, Van Den Bossche, Jbour, Ndang’Ang’a, Jones, Abdou, Al-Hmoud, Asswad, Atienza, Atrash, Barbara, Bensusan, Bino, Celada, Cherkaoui, Costa, Deceuninck, Etayeb, Feltrup-Azafzaf, Figelj, Gustin, Kmecl, Kocevski, Korbeti, Kotrošan, Laguna, Lattuada, Leităo, Lopes, López-Jiménez, Lucić, Micol, Moali, Perlman, Piludu, Portolou, Putilin, Quaintenne, Ramadan-Jaradi, Ružić, Sandor, Sarajli, Saveljić, Sheldon, Shialis, Tsiopelas, Vargas, Thompson, Brunner, Grimmett and Butchart2016). At the same time, conservation of wild bird species has been an important priority in Europe, as witnessed by the European Directive on birds (2009/147/EC), the first nature Directive adopted in 1979.
Cyprus is among the very few Mediterranean countries that have established a systematic monitoring scheme of illegal bird killing (BirdLife Cyprus 2015a, Brochet et al. Reference Brochet, Van Den Bossche, Jbour, Ndang’Ang’a, Jones, Abdou, Al-Hmoud, Asswad, Atienza, Atrash, Barbara, Bensusan, Bino, Celada, Cherkaoui, Costa, Deceuninck, Etayeb, Feltrup-Azafzaf, Figelj, Gustin, Kmecl, Kocevski, Korbeti, Kotrošan, Laguna, Lattuada, Leităo, Lopes, López-Jiménez, Lucić, Micol, Moali, Perlman, Piludu, Portolou, Putilin, Quaintenne, Ramadan-Jaradi, Ružić, Sandor, Sarajli, Saveljić, Sheldon, Shialis, Tsiopelas, Vargas, Thompson, Brunner, Grimmett and Butchart2016). Trapping, trading and consumption of small songbirds, although illegal, are still quite common on the island. Historical roots of this phenomenon go back to times when migratory birds offered much needed protein as a food supplement while more recently it also provided, due to the increase in scale, remarkable profits (Franzen Reference Franzen2010, Reference Franzen2013).
According to BirdLife Cyprus, around 400 bird species have been recorded in Cyprus, some 153 of which have been found among the illegally trapped birds. It is estimated that more than 2 million individuals are trapped illegally each year in Cyprus, and according to the recent review by Brochet et al. (Reference Brochet, Van Den Bossche, Jbour, Ndang’Ang’a, Jones, Abdou, Al-Hmoud, Asswad, Atienza, Atrash, Barbara, Bensusan, Bino, Celada, Cherkaoui, Costa, Deceuninck, Etayeb, Feltrup-Azafzaf, Figelj, Gustin, Kmecl, Kocevski, Korbeti, Kotrošan, Laguna, Lattuada, Leităo, Lopes, López-Jiménez, Lucić, Micol, Moali, Perlman, Piludu, Portolou, Putilin, Quaintenne, Ramadan-Jaradi, Ružić, Sandor, Sarajli, Saveljić, Sheldon, Shialis, Tsiopelas, Vargas, Thompson, Brunner, Grimmett and Butchart2016) the island holds by far the highest mean number of individual birds illegally harvested each year among Mediterranean countries (195.9 birds per 100 persons and 248.3 birds per km2 every year). Additionally, three of the top 20 trapping locations in the whole Mediterranean in terms of largest numbers of individuals being killed per year are found in Cyprus, the highest on the list (Famagusta area) being situated on the island (Brochet et al. Reference Brochet, Van Den Bossche, Jbour, Ndang’Ang’a, Jones, Abdou, Al-Hmoud, Asswad, Atienza, Atrash, Barbara, Bensusan, Bino, Celada, Cherkaoui, Costa, Deceuninck, Etayeb, Feltrup-Azafzaf, Figelj, Gustin, Kmecl, Kocevski, Korbeti, Kotrošan, Laguna, Lattuada, Leităo, Lopes, López-Jiménez, Lucić, Micol, Moali, Perlman, Piludu, Portolou, Putilin, Quaintenne, Ramadan-Jaradi, Ružić, Sandor, Sarajli, Saveljić, Sheldon, Shialis, Tsiopelas, Vargas, Thompson, Brunner, Grimmett and Butchart2016). Non-selective capture methods (lime-sticks, nets), in combination with the widely used tape luring, account for the highest number of species illegally caught. The main target species is the Blackcap Sylvia atricapilla; however, more than 40 species lump with the Blackcap under the collective term ambelopoulia (the local name for small migrating songbirds), such as the Lesser Whitethroat Sylvia curruca, Garden Warbler S. borin, various species of pipits, shrikes and Phylloscopus warblers, as well as the two endemic species (Cyprus Warbler S. melanothorax and Cyprus Wheatear Oenanthe cypriaca). Trapping of these species is illegal while the only ones that can be legally hunted are those listed in the transposition in Cyprus law of the Birds Directive 2009/147/EC, among the passerines, the Skylark Alauda arvensis, and some thrushes (Common Blackbird Turdus merula, Song Thrush T. philomelos, Redwing T. iliacus, and others).
Although there is no evidence that the main targeted species, S. atricapilla, is under direct threat, the non-selective practices that trappers use (mist-nets, lime-sticks) also trap many ‘by-catch’ species such as the congeneric S. conspicillata and S. melanothorax, many other passerines, as well as owls, nightjars, harriers, and falcons, several of which are of conservation concern. Considering that the abundance and biomass of common birds across Europe have undergone an unprecedented decline in the last 30 years (Inger et al. Reference Inger, Gregory, Duffy, Stott, Voříšek and Gaston2015), these capturing practices could pose a direct threat to bird biodiversity.
When enforcement authorities raid restaurants or perform searches in private houses, plucked birds cannot be identified as, apart from feathers, the head and legs are also usually removed. Hence, it is very difficult to argue in law courts about which species they are and whether they were acquired legally or not. In addition to its widespread application in taxonomy and biodiversity studies, DNA barcoding has proved to be an invaluable tool for researchers looking for an easy, quick and cost-effective diagnostic tool to identify species when morphological approaches fail (Hebert et al. Reference Hebert, Stoeckle, Zemlak and Francis2004, Frézal and Leblois Reference Frézal and Leblois2008). Considering the increasing number of DNA-based studies reliant on non-invasive sampling methods, the use of shed feathers, blood droplets or faecal droppings makes barcoding a potentially useful tool in many conservation fields (Rubinoff Reference Rubinoff2006, Francis et al. Reference Francis, Borisenko, Ivanova, Eger, Lim, Guillén-Servent, Kruskop, Mackie and Hebert2010). For instance, Cytochrome Oxidase subunit I (COI) mitochondrial DNA gene sequencing of endangered animals under poaching pressure allows low-cost and effective species identification, which can be essential for law enforcement and conservation officers (Cai et al. Reference Cai, Yue, Jiang, Xie, Li and Zhou2010).
Nevertheless, a series of concerns and criticisms were expressed after Hebert et al. (Reference Hebert, Cywinska and Ball2003) published their seminal paper on barcoding. Some authors challenged the use of universal genetic distance threshold values as a criterion to discriminate species (DeSalle et al. Reference DeSalle, Egan and Siddall2005, Wiemers and Fiedler Reference Wiemers and Fiedler2007). Others expressed worries concerning the underestimation of interspecific variation when closely related taxa are included in the analyses (Moritz and Cicero Reference Moritz and Cicero2004, Meyer and Paulay Reference Meyer and Paulay2005, Wiemers and Fiedler Reference Wiemers and Fiedler2007). Moreover, some authors claimed that insufficient sample size may compromise current results in more extensive future studies (Moritz and Cicero Reference Moritz and Cicero2004, Will et al. Reference Will, Mishler and Wheeler2005, Hickerson et al. Reference Hickerson, Meyer and Moritz2006).
These criticisms notwithstanding, the DNA barcoding approach has produced and still continues to produce, very useful data with several applications to conservation biology, ranging from anti-poaching, the fight against the illegal pet trade, and animal cruelty to the identification of bird species involved in aircraft strikes (see the review by Iyengar Reference Iyengar2014). DNA-based identification methods have been applied to a vast range of animal taxa: invertebrates (e.g. Weese and Santos Reference Weese and Santos2008), fish and/or fish products (e.g. Filonzi et al. Reference Filonzi, Chiesa, Vaghi and Nonnis Marzano2010, Doukakis et al. Reference Doukakis, Pikitch, Rothschild, DeSalle, Amato and Kolokotronis2012), reptiles (Gaur et al. Reference Gaur, Singh, Sreenivas and Singh2012, Welton et al. Reference Welton, Siler, Linkem, Diesmos, Diesmos, Sy and Brown2013), mammals (Fumagalli et al. Reference Fumagalli, Cabrita and Castella2009, Eaton et al. Reference Eaton, Meyers, Kolokotronis, Leslie, Martin and Amato2010, Dalton and Kotze Reference Dalton and Kotze2011, Barbanera et al. Reference Barbanera, Guerrini, Beccani, Forcina, Anayiotos and Panayides2012) and birds (Coghlan et al. Reference Coghlan, White, Parkinson, Haile, Spencer and Bunce2011, Abe et al. Reference Abe, Hayano and Inoue-Murayama2012, Aliabadian et al. Reference Aliabadian, Beentjes, Roselaar (Kees), van Brandwijk, Nijman and Vonk2013). Among these approaches, those relying on DNA barcoding have provided a significant contribution, taking advantage of the opportunity to identify species from deformed, destroyed, cooked or otherwise processed tissues, even in traces (Weese and Santos Reference Weese and Santos2008, Eaton et al. Reference Eaton, Meyers, Kolokotronis, Leslie, Martin and Amato2010, Filonzi et al. Reference Filonzi, Chiesa, Vaghi and Nonnis Marzano2010, Coghlan et al. Reference Coghlan, White, Parkinson, Haile, Spencer and Bunce2011, Dalton and Kotze Reference Dalton and Kotze2011, Abe et al. Reference Abe, Hayano and Inoue-Murayama2012, Aliabadian et al. Reference Aliabadian, Beentjes, Roselaar (Kees), van Brandwijk, Nijman and Vonk2013, Welton et al. Reference Welton, Siler, Linkem, Diesmos, Diesmos, Sy and Brown2013). In Cyprus, for example, it is well-known that trapped migrating birds are preserved in vinegar and/or cooked before they are served illegally in restaurants. DNA-based methods have already been applied to protect Cypriot wildlife in a forensic context, such as in the case of poaching of the endemic mouflon Ovis orientalis ophion (Barbanera et al. Reference Barbanera, Guerrini, Beccani, Forcina, Anayiotos and Panayides2012).
This work aimed to create a COI sequence database of all bird species resident in Cyprus, as well as those most commonly trapped (both targeted and common by-catch species). The database could be used to identify confiscated or otherwise collected samples of tissue or whole birds, even when these samples have been processed (e.g. cooked), badly preserved or are not morphologically identifiable. Even though COI sequence data are already available for many species included in our study, these come from populations that mostly belong to other geographic regions. Hence, available DNA sequences might not be appropriate for identification of resident species in an isolated island such as Cyprus. For instance, it is worth noting that Cyprus hosts an endemic species (Cyprus Warbler) that is congeneric with target ambelopoulia species (Blackcap). Therefore, the use of samples from local populations was considered essential for forensic identification.
Materials and methods
Sampling in the wild (between 2012 and 2015) was carried out by BirdLife Cyprus and the Game and Fauna Service, the public authority in charge of all issues related to hunting, poaching and illegal trapping in Cyprus. BirdLife Cyprus collected samples (feathers) mostly during routine ringing activity, while the Game and Fauna Service obtained samples from confiscations of unplucked birds, from hunters as well as from injured or exhausted birds treated in the Service’s wildlife rehabilitation centre. Whenever possible, samples from road-killed birds were also collected. Furthermore, other types of tissues such as dry blood on filter paper, skin and muscle taken from dead individuals were utilised. Overall, 103 samples belonging to 81 species were collected and sequenced (Table S1 in the online supplementary material).
All individuals were identified on the basis of morphological characters and usually 1-2 feathers were collected from trapped birds before they were released. The vast majority of samples were preserved in 96% ethanol and stored at -20o C. One sample of domestic chicken was cooked (boiled at 100o C for 90 min), and four samples (three Blackcaps and one Chukar Partridge Alectoris chukar) were preserved in vinegar for a minimum of 12 months, with the Blackcaps being also subsequently fried.
DNA extraction
Genomic DNA was extracted from 103 individuals representing 81 different taxa (Table S1). Two different commercial DNA extraction kits, namely the Dneasy Blood and Tissue kit (Qiagen) and the Nucleo Spin Food kit (Macherey-Nagel) were used, following the manufacturer’s instructions. The latter was specifically employed in the event of either cooked-meat or not well-preserved sample.
DNA amplification and sequencing
DNA concentration was determined for each sample using NanoDrop 2000/200c (Thermo Fisher Scientific Inc., USA). All PCR reactions were adjusted to a final volume of 40 μl. Each reaction contained 0.2 µL of Kapa Taq DNA Polymerase (5U/μL), 2.4 μL of 25 mM MgCl2, 4 μL of Kapa 10X PCR buffer A, 1.2 μL of 10mM dNTP (Kapa Biosystems, Inc., UK), 1.2 μL of each primer (10 µM) and c.20 ng of DNA template. Reactions were performed in a Veriti thermal cycler (Applied Biosystems, USA) with the following thermal profile: 3 min at 94° C (initial denaturation); 1 min at 94° C, 1 min at [47-50-55]° C (depending on the primer pair used, Table 1), and 1 min at 72° C (39 cycles); 7 min at 72° C (final extension). The amplification of the targeted fragment (COI, 648bp) was achieved using three primer pairs (Table 1). For all samples, first PCR reactions were carried out using the L6615(tTyr) COI/H7548 (COI) primer pair. When these primers failed to amplify the targeted gene, the next primer pair used was LCO1490/HCO2198 and then BirdF1/COIbirdR2. These two latter pairs and a combination of these with L6615(tTyr) COI/H7548 (COI) were also used in nested and semi-nested PCR (Guerrini and Barbanera Reference Guerrini and Barbanera2009), respectively, using the first PCR reaction product as template. Nested and semi-nested PCR were employed mostly in cases where samples were processed (i.e. cooked, samples in vinegar) or not well-preserved (i.e. shed feathers). Detailed information on PCR procedures followed is given in the supplementary material.
Table 1. Code, sequence, product size, annealing temperature and literature records of primer pairs used in this study.

PCR products were purified with the Qiaquick Purification Kit (Qiagen, Germany) following the manufacturer’s instructions. Purified products were directly sequenced at Macrogen (Amsterdam, The Netherlands) on both DNA strands. One additional sequence of domestic chicken obtained from GenBank, was included in analyses and designed as an outgroup in the phylogenetic reconstructions. GenBank accession numbers of all sequences produced in the present work were provided in Table S1.
Data elaboration and phylogenetic analyses
Sequences were edited with CodonCode Aligner (v. 3.7.1; CodonCode Corp., USA), which was also used to assemble contigs. Alignment was performed using ClustalX (Thompson et al. Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997) while MEGA v.6 (Tamura et al. Reference Tamura, Stecher, Peterson, Filipski and Kumar2013) was used to calculate corrected pairwise genetic distances under the TN93 model of evolution (Tamura and Nei Reference Tamura and Nei1993). The latter, indeed, accounts for unequal base frequency and different rates for transitions (Ti) and transversions (Tv) typical for mtDNA. Haplotypes (H) were inferred by means of DnaSP v. 10.1 (Librado and Rozas Reference Librado and Rozas2009).
To assess the mitochondrial authenticity of all PCR products, COI sequences were carefully inspected in order to check for the possible contamination from Numts (mitochondrial sequences of nuclear origin: Lopez et al. Reference Lopez, Yuhki, Masuda, Modi and O’Brien1994) according to the following criteria: (i) lack of internal stop codons and/or gaps; (ii) expected G-biased nucleotide frequency at 3rd codon position (typical of animal mtDNA); (iii) expected Ti/Tv ratio larger than 1; (iv) unambiguous alignment of retrieved sequences and their correspondence at conserved nucleotide positions with available GenBank sequences (Sorenson and Quinn Reference Sorenson and Quinn1998, Sheldon et al. Reference Sheldon, Jones and McCracken2000).
Sequences were tested in order to check whether they conformed to a neutral model of evolution as expected for mtDNA (Tajima Reference Tajima1989; test as implemented in DnaSP). Phylogenetic signal was evaluated by means of the index of substitution saturation (Iss) using Xia’s test with 1,000 replicates (Xia et al. Reference Xia, Xie, Salemi, Chen and Wang2003) as implemented in DAMBE v. 5.6.9 (Xia Reference Xia2013), and by plotting the number of Ti and Tv against the TN93-corrected genetic distance. TN93 evolutionary model is the best among the few available in DAMBE, which to our knowledge is the only software where Xia’s test can be performed. This was another reason for selecting this model, which, additionally, is also formally suitable for the analysis.
The best DNA substitution model fitting our data set was selected according to the Akaike’s Information Criterion (AIC; Akaike Reference Akaike1974) using jModelTest v.2.1.1 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012). Likelihood scores were calculated according to the following settings: three substitution schemes, base frequencies estimation, gamma shape (four categories) and invariable sites estimation. Models including both gamma distribution and invariable sites were neglected (Yang, 2006). Out of 88 candidate models, the General Time Reversible (GTR; Rodríguez et al. Reference Rodríguez, Oliver, Marín and Medina1990) + gamma (G) was chosen with a log likelihood - In = 15220.8468 (α = 0.171).
Phylogenetic trees were constructed using Bayesian Inference (BI) and Maximum Likelihood (ML) methods. Estimated parameters were applied to the Bayesian analysis conducted with Metropolis-coupled Markov chain Monte Carlo algorithms as implemented in MrBayes (v. 3.1.2; Ronquist and Huelsenbeck Reference Ronquist and Huelsenbeck2003). Four independent analysis runs (eight chains per run, each starting from independent random trees) were conducted for 3,000,000 generations with a sample frequency of 100. Convergence among runs was monitored in MrBayes through the average standard deviation of split frequencies, and runs were continued until this value dropped well under 0.01. Then, the convergence of each run towards stationarity was monitored with Tracer (v. 1.5; Rambaut and Drummond Reference Rambaut and Drummond2007) using likelihood values as well as all other parameters estimated, and stationarity was reached after 750,000 generations. Hence, 30,000 trees were discarded as burn-in, and the remaining 90,004 trees were used to produce 50% majority-rule consensus trees.
Maximum Likelihood reconstruction was carried out using online PhyML v. 3.0 platform (Guindon et al. Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) platform with Nearest-Neighbor-Interchange search, Bio-Neighbor Joining starting tree under the GTR evolutionary model. Reliability at each node of the ML tree was assessed by bootstrapping (Felsenstein Reference Felsenstein1985) with 1,000 replicates.
Results
No internal stop codons or gaps were detected: Ti/Tv ratio was 1.188, and the G frequency at the 3rd codon position was 4.6%. Sequences conformed to a neutral model of evolution (Tajima’s D = -0.032, P > 0.05). The analysis of the phylogenetic signal (outgroup included) did not show any significant level of saturation. Xia’s test provided an Iss value (= 0.214) that was significantly smaller (P < 0.001) than that of the critical Iss (Iss.c, the Iss value at which the sequences begin to fail to recover the true tree) assuming either symmetrical or asymmetrical topology for the phylogenetic reconstructions (Iss.cSym = 0.717 and Iss.cAsym = 0.391, respectively). In the plot of the number of the Ti and Tv versus divergence measured by the TN93 evolutionary model (Figure S1 in the supplementary material), the Ti/Tv ratio was fairly larger than 1, some sign of saturation showing up only for genetic distance values larger than 23%.
Aligned sequences (all 648 bp-long, reading frame = 1) matched with nucleotide positions starting from site 6714 to site 7361 of G. gallus given by Nishibori et al. (Reference Nishibori, Hanazono, Yamamoto, Tsudzuki and Yasue2003) with GenBank acc.no.: AP003317.1. Ninety-two haplotypes obtained from 103 sampled individuals accounted for a total of 81 taxa (Table 1). Among the aligned 648 nucleotide sites, 284 were variable and 265 were parsimony informative. Interspecific pairwise genetic distances ranged between 0.3% (Common Buzzard Buteo buteo versus Long-legged Buzzard B. rufinus) and 25.1% (Short-toed Treecreeper Certhia brachydactyla, versus Corncrake Crex crex). Unique polymorphisms between congeneric species of Sylvia were also detected (Table 2).
Table 2. Diagnostic polymorphisms of Blackcap S. atricapilla, compared with other Sylvia species living in Cyprus.
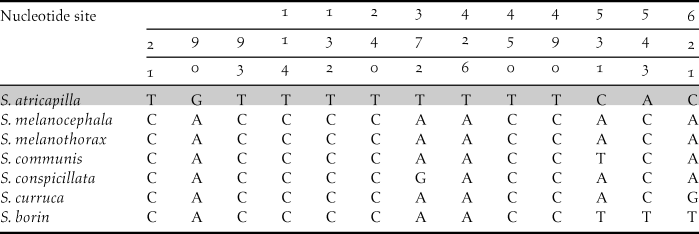
Finally, six Cypriot COI sequences were new entries to the GenBank, i.e., those of Bonelli’s Eagle Aquila fasciata, Long-legged buzzard, Eleonora’s Falcon Falco eleonorae, Spectacled Warbler Sylvia conspicillata, Cyprus Warbler and Masked Shrike Lanius nubicus.
BI and ML phylogenetic tree reconstructions carried out under the same evolutionary model produced overlapping topologies, hence, only the 50% majority-rule consensus BI tree is given in Figure 1.

Figure 1. Phylogenetic reconstruction obtained by Bayesian Inference using Gallus gallus (haplotypes H1 and H2) as outgroup. Labels correspond to haplotypes given in Table S1. Maximum Likelihood (ML) reconstructions provided a largely overlapping topology. Hence, the statistic support was reported at each node as it follows: left, posterior probability values (> 0.90) computed in the BI analysis; right, bootstrap percentage values (> 50) computed in the ML tree.
Discussion
Recently developed genetic tools, such as DNA barcoding, are increasingly used within conservation biology (Iyengar Reference Iyengar2014). Numerous studies incorporate genetic data to fight poaching and smuggling, assist an effective application of CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora), identify samples of forensic interest, trace food, etc. (Dalton and Kotze Reference Dalton and Kotze2011, Abe et al. Reference Abe, Hayano and Inoue-Murayama2012, Barbanera et al. Reference Barbanera, Guerrini, Beccani, Forcina, Anayiotos and Panayides2012). To further contribute to this endeavour, the DNA barcoding of avian species resident in Cyprus and several common migrants which are potential victims of illegal trapping was successfully accomplished. Hence, a DNA database enabling reliable, fast and cost-effective identification of bird samples was made available.
Overall, DNA barcodes were generated for 14 legally hunted bird species (including six that are sometimes served in restaurants), 43 species among those illegally trapped and locally referred to as ambelopoulia plus 23 by-catch species (not including domestic chicken; see Table S1). Furthermore, the assignment of genetic ID to boiled and fried meat or meat stored in vinegar proved that a valuable support to law enforcement authorities is now possible following sample confiscations in restaurants, houses or other premises. Therefore, from now on the identification of plucked or cooked birds through DNA barcoding can assist prosecution of poachers and ambelopoulia trappers.
In the present study, DNA was successfully amplified from ill-preserved samples such as shed feathers, biological traces (blood), even human-processed tissue (cooked or preserved in vinegar) by using a combination of nested and semi-nested PCR with different primer pairs. This result is very important considering that DNA investigation of similarly processed bird specimens will be demanded often, this being the most common way of preserving and serving small songbirds in Cyprus. Consequently, the protocols followed and the dataset produced herein will provide the necessary background for future species identification of various and differently preserved tissues. The results of this work, as well as other published studies with larger datasets, show that specimen identification through DNA barcoding is effective and reliable, even when they belong to sister taxa. Indeed, extremely low intraspecific genetic distances at the selected region in contrast with relatively high interspecific distances among congeneric species allow for safe identification (e.g. Tavares and Baker Reference Tavares and Baker2008, Ward Reference Ward2009).
However, DNA barcoding cannot always provide reliable specimen identification, such as in the case of taxa with intra- and inter-specific genetic distance range overlapping to a significant extent. Nevertheless, it has been shown that most of these cases were primarily due to wrong morphological identification of specimens carried out before genetic analysis (e.g. Wiemers and Fiedler Reference Wiemers and Fiedler2007, Tavares and Baker Reference Tavares and Baker2008, Ward Reference Ward2009, Tavares et al. Reference Tavares, Gonçalves, Miyaki and Baker2011). In the case of Cypriot birds, we demonstrate DNA barcoding to be an effective and reliable tool able to assign genetic ID to all species investigated. Another important contribution we present is the disclosure of diagnostic nucleotide polymorphisms within the genus Sylvia, including a target species of conservation concern, Blackcap, present in both the newly produced sequences and those retrieved from GenBank (Table 2). This finding provided additional conservation value to the dataset produced herein, as an easily recognizable barcode is now available for a group of taxa whose genetic distances might not provide otherwise safe identification (Tavares and Baker Reference Tavares and Baker2008).
Even though a robust and fully resolved phylogeny of birds based solely on one mtDNA gene fragment was not expected, the results of this study may also contribute to avian phylogenetic investigation, as shown by the large amount of previously established, well-supported clades recovered in the tree (Figure 1). Our phylogenetic reconstructions provided weight against published criticisms on the effectiveness of DNA-barcoding when based on a single gene fragment and dealing with closely related taxa (e.g. Moritz and Cicero Reference Moritz and Cicero2004, Meyer and Paulay Reference Meyer and Paulay2005, Wiemers and Fiedler Reference Wiemers and Fiedler2007). In fact, the phylogenetic reconstruction in Figure 1 is in large agreement with known bird phylogeny (Jetz et al. Reference Jetz, Thomas, Joy, Hartmann and Mooers2012, Prum et al. Reference Prum, Berv, Dornburg, Field, Townsend, Lemmon and Lemmon2015), all named species are identified as separate taxa, and the monophyly for almost all genera including more than one species has been recovered as well as many sister-clade relationships at the genus level. Nevertheless, some misplacements (e.g. Tyto alba between falcons and coots) and discrepancies (e.g. doves as sister-clade to eagles and buzzards) occurred as well. In any case, even though the phylogenetic tree is based on a single gene and cannot be considered as a robust estimate of phylogenetic relationships, it still highlights the adequacy of COI to discriminate among taxa and establish reasonable relationships among target species. Hence it can be used effectively for the intended conservation purposes.
Application of our experimental protocol and DNA barcoding database provides a tool to compare wildlife forensic evidence, enabling Cypriot authorities to prove guilt regardless of whether suspects were caught in the act or not. Until now, law enforcement authorities in Cyprus have not used DNA evidence against illegal bird trapping especially because sequences available for the species of interest were obtained from individuals from other geographic regions. Far beyond direct punishment of offenders, tools such as DNA barcoding allow also for the reduction of efforts needed to collect poaching evidence in the field. This, in turn, may result in a much lower risk for police officers or Game and Fauna Service personnel integrity, as genetic evidence is objective beyond any in situ dispute. Furthermore, the ability to identify processed or cooked birds can strongly contribute to the reduction of poaching as income, since restaurants are an important component of the illegal bird market. According to existing laws, restaurant owners who illegally serve birds are considered guilty as are the trappers themselves. DNA-based specimen identification will allow for more effective investigation of local restaurant owners. Thus, the use of DNA barcoding as a forensic investigative tool may represent an effective deterrent for restaurant owners or other individuals that possess protected species.
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0959270916000472
Acknowledgements
This research was funded by the A.G. Leventis Foundation and the Tasso Leventis Conservation Foundation. The authors would like to thank BirdLife Cyprus staff and volunteers who collected dead birds they found during the sampling period and participated in ringing activities. The authors are deeply grateful to Michalis Antoniou and Charalambos Hadjistyllis (Officers at the Game and Fauna Service, Ministry of Interior, Cyprus) for sample collection as well as for their training at the University of Pisa on species identification through DNA barcoding, an experimental background that will assist their work in future cases involving people under prosecution. Finally, we would like to thank two anonymous reviewers and Edwin Harris, Associate Editor, for their valuable comments on a previous draft of this manuscript that greatly improved the quality of this text.







